-Search query
-Search result
Showing 1 - 50 of 55 items for (author: hofmann & h)

EMDB-14156: 
Cryo-EM reconstruction of the Bacillus subtilis MutS2-collided disome complex (MutS2 conf.1; Leading 70S)
Method: single particle / : Filbeck S, Pfeffer S

EMDB-14157: 
Composite reconstruction of the Bacillus subtilis collided disome (Leading 70S)
Method: single particle / : Filbeck S, Pfeffer S

EMDB-14158: 
Composite reconstruction of the Bacillus subtilis collided disome (Collided 70S)
Method: single particle / : Filbeck S, Pfeffer S

EMDB-14159: 
Cryo-EM reconstruction of the Bacillus subtilis MutS2-collided disome complex (MutS2 conf.2; Leading 70S)
Method: single particle / : Filbeck S, Pfeffer S

EMDB-14160: 
Cryo-EM reconstruction of the Bacillus subtilis MutS2-collided disome complex (Leading 70S)
Method: single particle / : Filbeck S, Pfeffer S

EMDB-14161: 
Cryo-EM reconstruction of the Bacillus subtilis MutS2-collided disome complex (Collided 70S)
Method: single particle / : Filbeck S, Pfeffer S

EMDB-14162: 
Cryo-EM reconstruction of the Bacillus subtilis collided disome (Leading 70S)
Method: single particle / : Filbeck S, Pfeffer S

EMDB-14163: 
Cryo-EM reconstruction of the Bacillus subtilis collided disome (Leading 30S)
Method: single particle / : Filbeck S, Pfeffer S

EMDB-14164: 
Cryo-EM reconstruction of the Bacillus subtilis collided disome (Collided 70S)
Method: single particle / : Filbeck S, Pfeffer S

EMDB-14165: 
Cryo-EM reconstruction of the Bacillus subtilis collided disome (Collided 30S)
Method: single particle / : Filbeck S, Pfeffer S

EMDB-14166: 
Cryo-EM reconstruction of Bacillus subtilis obstructed 50S subunit co-purified with MutS2
Method: single particle / : Filbeck S, Pfeffer S

PDB-7qv1: 
Bacillus subtilis collided disome (Leading 70S)
Method: single particle / : Filbeck S, Pfeffer S

PDB-7qv2: 
Bacillus subtilis collided disome (Collided 70S)
Method: single particle / : Filbeck S, Pfeffer S

PDB-7qv3: 
Bacillus subtilis MutS2-collided disome complex (MutS2 conf.2; Leading 70S)
Method: single particle / : Filbeck S, Pfeffer S

EMDB-11022: 
Allostery through DNA drives phenotype switching
Method: single particle / : Rosenblum G, Elad N, Rozenberg H, Wiggers F, Jungwirth J, Hofmann H

EMDB-12260: 
Allostery through DNA drives phenotype switching
Method: single particle / : Rosenblum G, Elad N

PDB-6z0s: 
Allostery through DNA drives phenotype switching
Method: single particle / : Rosenblum G, Elad N, Rozenberg H, Wiggers F, Jungwirth J, Hofmann H

PDB-7nbn: 
Allostery through DNA drives phenotype switching
Method: single particle / : Rosenblum G, Elad N, Rozenberg H, Wiggers F, Jungwirth J, Hofmann H
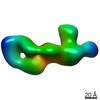
EMDB-20268: 
Chikungunya virus p62/E1 complex
Method: single particle / : Pallesen J, Ward AB

EMDB-20269: 
Chikungunya virus p62/E1 in complex with Fab from IgG DC2.271B
Method: single particle / : Pallesen J, Ward AB

EMDB-4773: 
Heterodimeric ABC exporter TmrAB in inward-facing narrow conformation under turnover conditions
Method: single particle / : Januliene D, Hofmann S, Medhdipour AR, Thomas C, Hummer G, Tampe R, Moeller A

EMDB-4774: 
Heterodimeric ABC exporter TmrAB in inward-facing wide conformation under turnover conditions
Method: single particle / : Januliene D, Hofmann S, Medhdipour AR, Thomas C, Hummer G, Tampe R, Moeller A

EMDB-4775: 
Heterodimeric ABC exporter TmrAB in ATP-bound outward-facing open conformation
Method: single particle / : Januliene D, Hofmann S, Medhdipour AR, Thomas C, Hummer G, Tampe R, Moeller A

EMDB-4776: 
Heterodimeric ABC exporter TmrAB in ATP-bound outward-facing occluded conformation
Method: single particle / : Januliene D, Hofmann S, Medhdipour AR, Thomas C, Hummer G, Tampe R, Moeller A

EMDB-4777: 
Heterodimeric ABC exporter TmrAB in vanadate trapped outward-facing open conformation
Method: single particle / : Januliene D, Hofmann S, Medhdipour AR, Thomas C, Hummer G, Tampe R, Moeller A

EMDB-4778: 
Heterodimeric ABC exporter TmrAB in vanadate trapped outward-facing occluded conformation
Method: single particle / : Januliene D, Hofmann S, Medhdipour AR, Thomas C, Hummer G, Tampe R, Moeller A

EMDB-4779: 
Heterodimeric ABC exporter TmrAB under turnover conditions in asymmetric unlocked return conformation
Method: single particle / : Januliene D, Hofmann S, Medhdipour AR, Thomas C, Hummer G, Tampe R, Moeller A

EMDB-4780: 
Heterodimeric ABC exporter TmrAB under turnover conditions in asymmetric unlocked return conformation with wider opened intracellular gate
Method: single particle / : Januliene D, Hofmann S, Medhdipour AR, Thomas C, Hummer G, Tampe R, Moeller A

EMDB-4781: 
Heterodimeric ABC exporter TmrAB with bound peptide substrate in inward-facing wide conformation
Method: single particle / : Januliene D, Hofmann S, Medhdipour AR, Thomas C, Hummer G, Tampe R, Moeller A

PDB-6raf: 
Heterodimeric ABC exporter TmrAB in inward-facing narrow conformation under turnover conditions
Method: single particle / : Thomas C, Januliene D, Mehdipour AR, Hofmann S, Hummer G, Moeller A, Tampe R

PDB-6rag: 
Heterodimeric ABC exporter TmrAB in inward-facing wide conformation under turnover conditions
Method: single particle / : Thomas C, Januliene D, Mehdipour AR, Hofmann S, Hummer G, Moeller A, Tampe R

PDB-6rah: 
Heterodimeric ABC exporter TmrAB in ATP-bound outward-facing open conformation
Method: single particle / : Thomas C, Januliene D, Mehdipour AR, Hofmann S, Hummer G, Moeller A, Tampe R

PDB-6rai: 
Heterodimeric ABC exporter TmrAB in ATP-bound outward-facing occluded conformation
Method: single particle / : Thomas C, Januliene D, Mehdipour AR, Hofmann S, Hummer G, Moeller A, Tampe R

PDB-6raj: 
Heterodimeric ABC exporter TmrAB in vanadate trapped outward-facing open conformation
Method: single particle / : Thomas C, Januliene D, Mehdipour AR, Hofmann S, Hummer G, Moeller A, Tampe R

PDB-6rak: 
Heterodimeric ABC exporter TmrAB in vanadate trapped outward-facing occluded conformation
Method: single particle / : Thomas C, Januliene D, Mehdipour AR, Hofmann S, Hummer G, Moeller A, Tampe R

PDB-6ral: 
Heterodimeric ABC exporter TmrAB under turnover conditions in asymmetric unlocked return conformation
Method: single particle / : Thomas C, Januliene D, Mehdipour AR, Hofmann S, Hummer G, Moeller A, Tampe R

PDB-6ram: 
Heterodimeric ABC exporter TmrAB under turnover conditions in asymmetric unlocked return conformation with wider opened intracellular gate
Method: single particle / : Thomas C, Januliene D, Mehdipour AR, Hofmann S, Hummer G, Moeller A, Tampe R

PDB-6ran: 
Heterodimeric ABC exporter TmrAB in inward-facing wide conformation
Method: single particle / : Thomas C, Januliene D, Mehdipour AR, Hofmann S, Hummer G, Moeller A, Tampe R
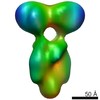
EMDB-0102: 
symmetric structure of tPDE6
Method: single particle / : Qureshi BM, Behrmann E, Loerke J, Spahn CMT, Heck M

EMDB-7786: 
human PKD2 F604P mutant
Method: single particle / : Zheng W, Yang X, Bulkley D, Chen XZ, Cao E

PDB-6d1w: 
human PKD2 F604P mutant
Method: single particle / : Zheng W, Yang X, Bulkley D, Chen XZ, Cao E

EMDB-3906: 
Cryo-EM density map of the human PLC editing module
Method: single particle / : Januliene D, Blees A, Trowitzsch S, Tampe R, Moeller A

PDB-6eny: 
Structure of the human PLC editing module
Method: single particle / : Trowitzsch S, Januliene D, Blees A, Moeller A, Tampe R

EMDB-3904: 
CryoEM density map of the pseudosymmetric PLC editing modules
Method: single particle / : Januliene D, Blees A, Trowitzsch S, Tampe R, Moeller A

EMDB-3905: 
Cryo-EM density map of the human MHC-I peptide loading complex
Method: single particle / : Januliene D, Blees A, Trowitzsch S, Tampe R, Moeller A

EMDB-2031: 
Unsymmetrized reconstruction of the FLNa16-21 rod2 segment
Method: single particle / : Hofmann GW, Jiang P, Campbell ID, Gilbert RJC

EMDB-2032: 
Twofold symmetrized reconstruction of the FLNa16-21 rod2 segment
Method: single particle / : Hofmann GW, Jiang P, Campbell ID, Gilbert RJC
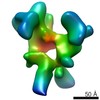
EMDB-2033: 
Unsymmetrized reconstruction of the FLNa16-21 rod2 segment bound with integrin Beta7 subunit tail
Method: single particle / : Hofmann GW, Jiang P, Campbell ID, Gilbert RJC
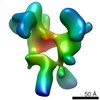
EMDB-2034: 
Twofold symmetrized reconstruction of the FLNa16-21 rod2 segment bound with integrin Beta7 subunit tail
Method: single particle / : Hofmann GW, Jiang P, Campbell ID, Gilbert RJC

EMDB-1328: 
Reconfiguration of yeast 40S ribosomal subunit domains by the translation initiation multifactor complex.
Method: single particle / : Gilbert RJC, Gordiyenko Y, von der Haar T
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
